
Tutorial
In this short tutorial a set of 1141475 HIV integration sites are analysed using Enhort.
Upload integration sites
At first, integration sites have to uploaded in a .bed-file. In this example set of about 1m HIV integration sites taken from the Retrovirus Integration Database is used. The wizard is selected on the main page:
And on the following page the .bed-file is selected on the local computer.
Select annotations
The sites are then uploaded and the overview for available annotations is shown:
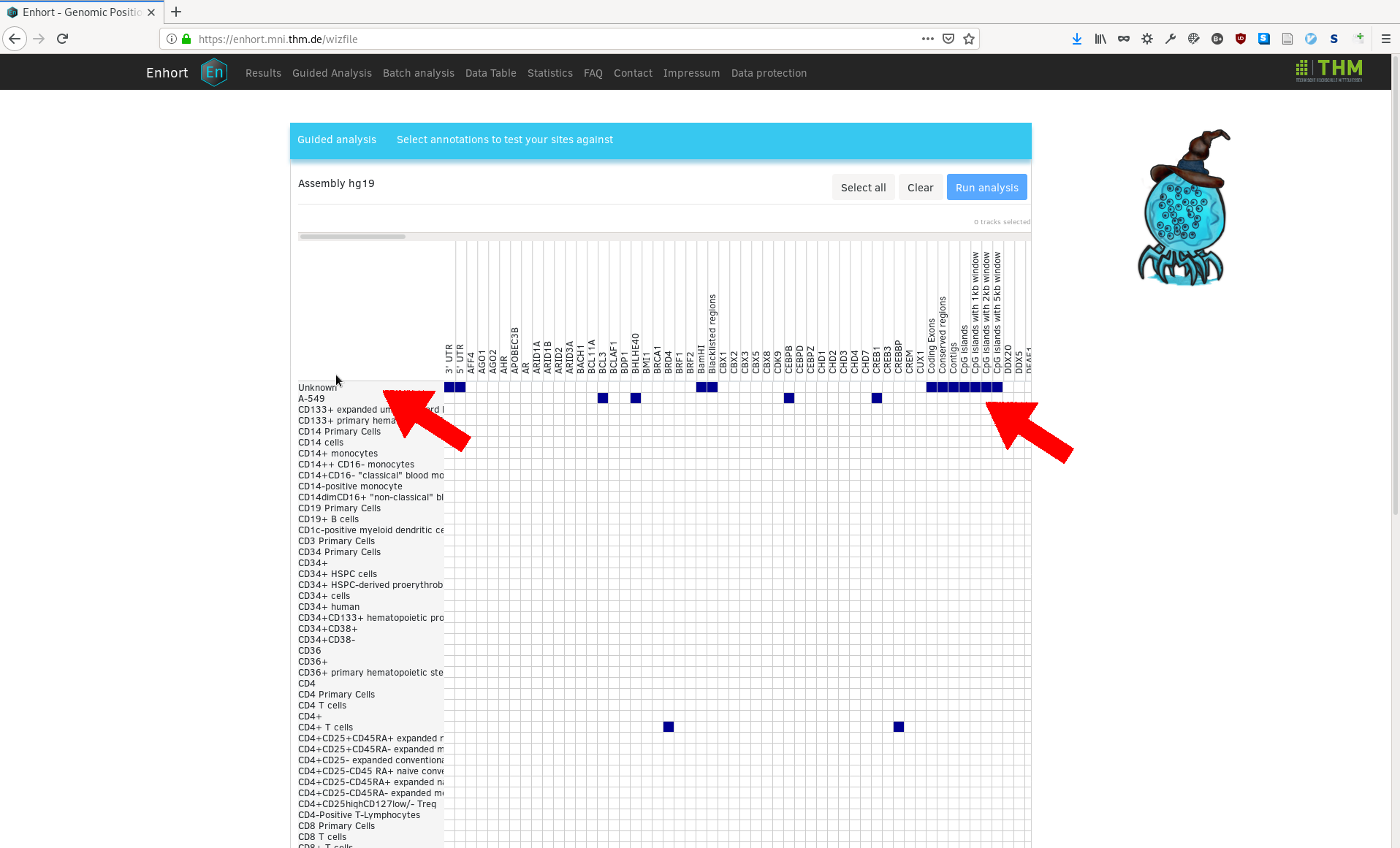
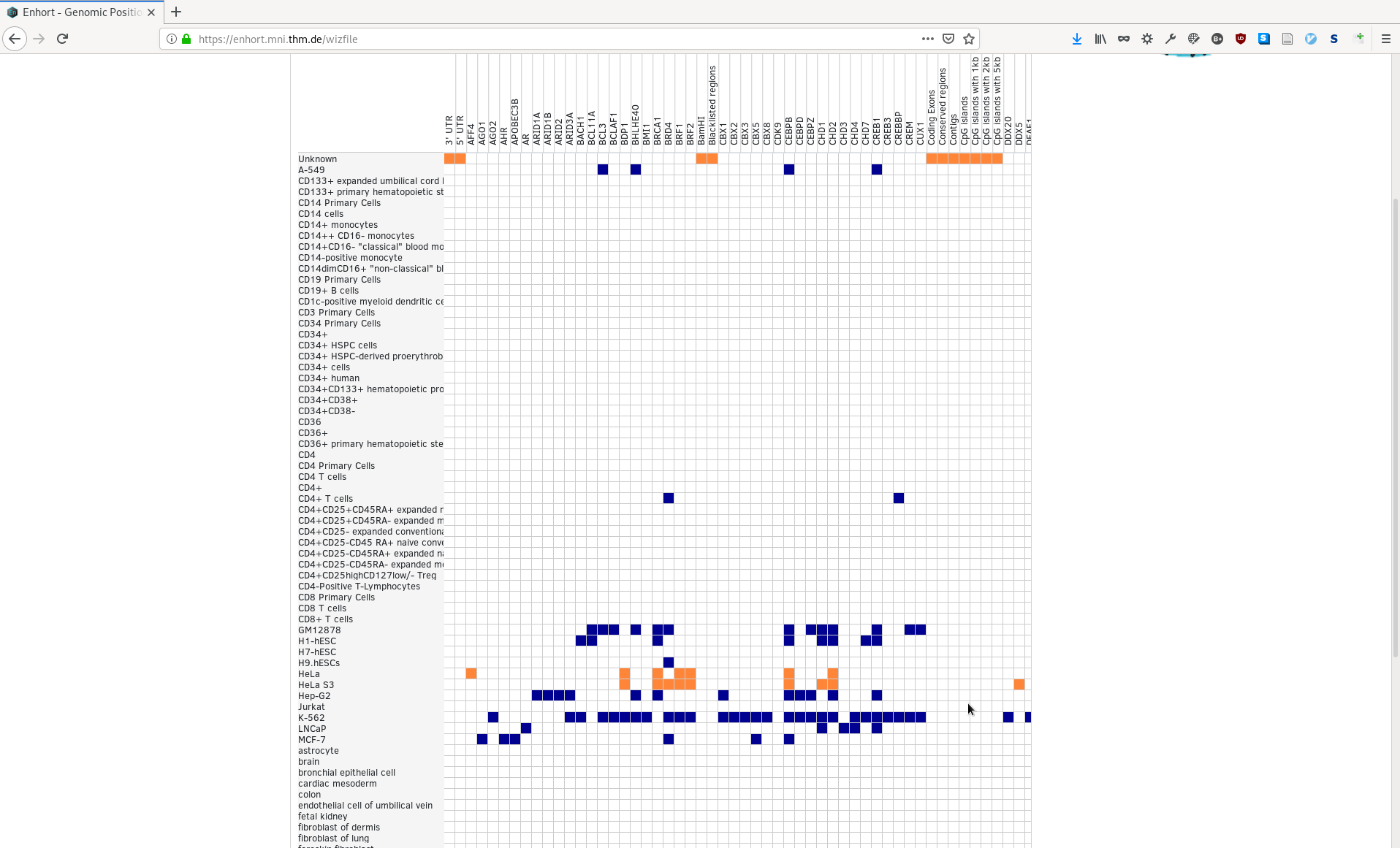
Single annotations can be selected by clicking on the blue squares. A set of annotations for a cell line or all annotations in a column can be selected by clicking on the row or column header. For example, all cell line independent annotations are selected in the following example, as well as annotations for HeLa:
Review results and add a covariate
The results for the given sites and selected covariates are then displayed in a table, containing the effect size and p value of each annotation, as well as overview over the annotation groups. Each row can be expanded to get exact integration counts for each annotation:
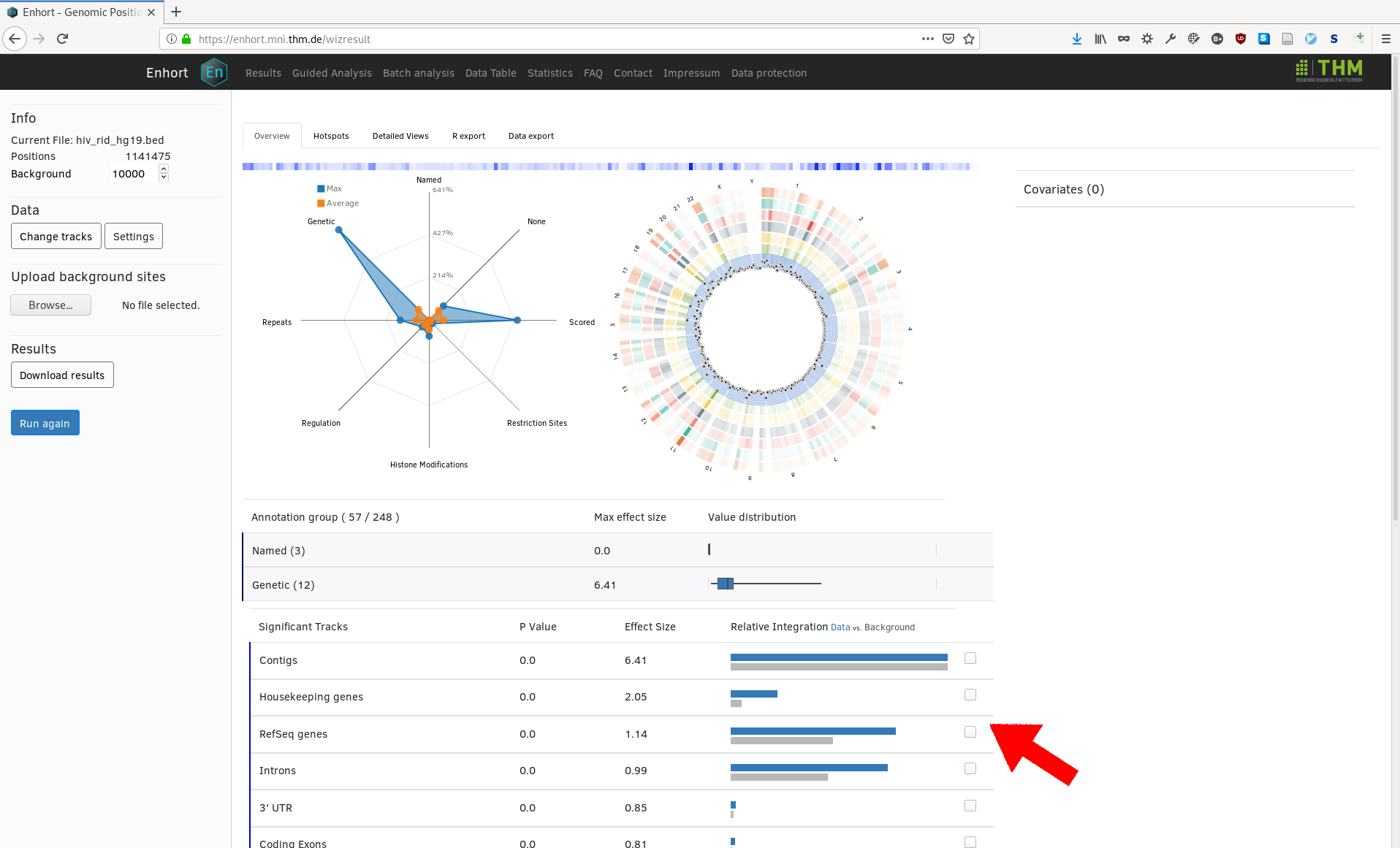
From here the user can select covariates to alter the integation preference of the background control sites. For example, HIV is known to integrate near genes which is observable in the upper image. The annotation track RefSeq genes can be selected as covariate to adapt the background for this known preference. To do this the marked checkbox is selected and with the button 'Run again' the background is changed. In the following image we see RefSeq genes was added to list of covariate on the right and now has the exact same integration preference as the HIV sites:
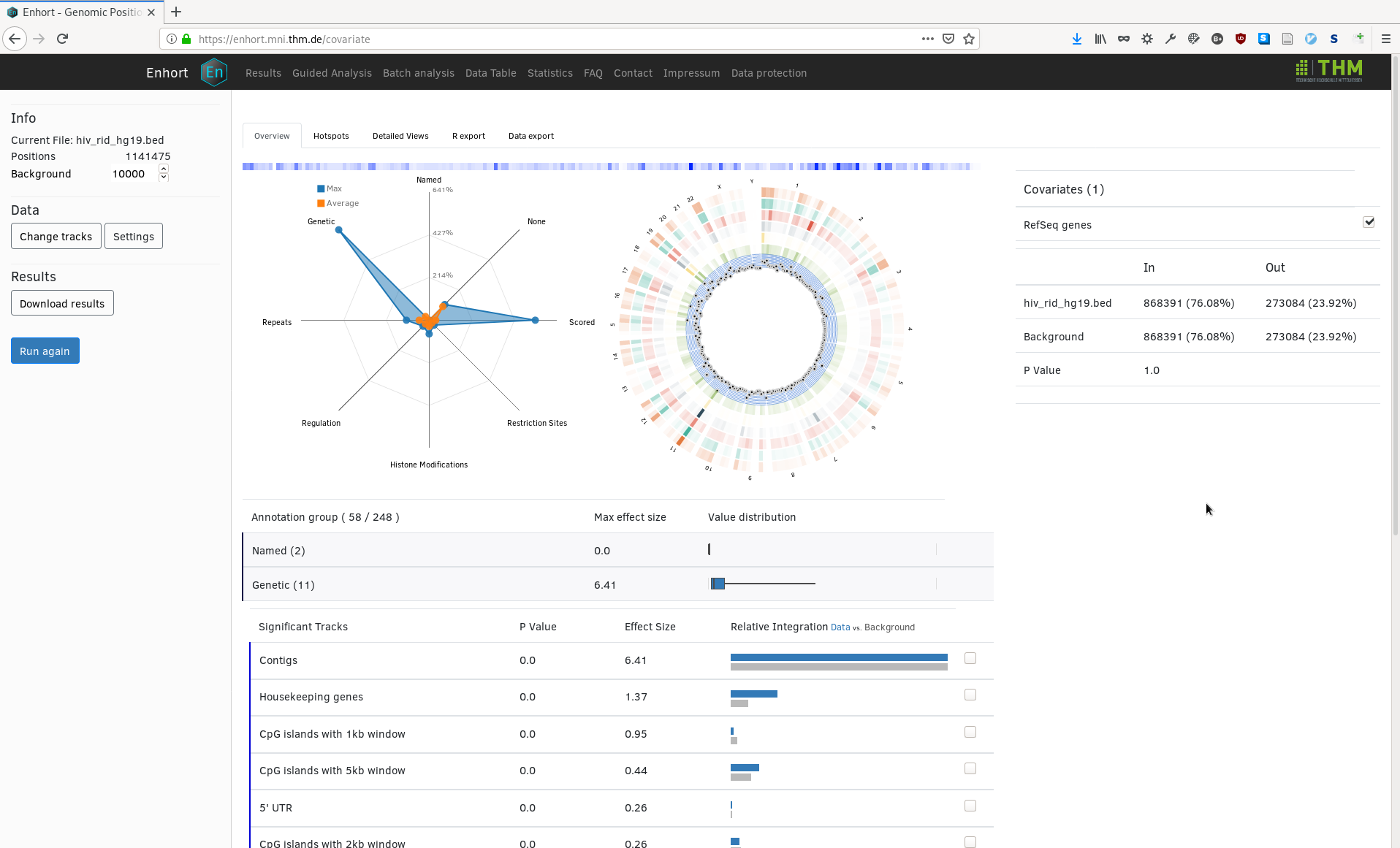
This adaptation then changes the significance and fold change of several annotations tracks in the main table. In the following the complete table of selected annotations is shown with 'houskeeping genes' expanded to show exact integration counts:
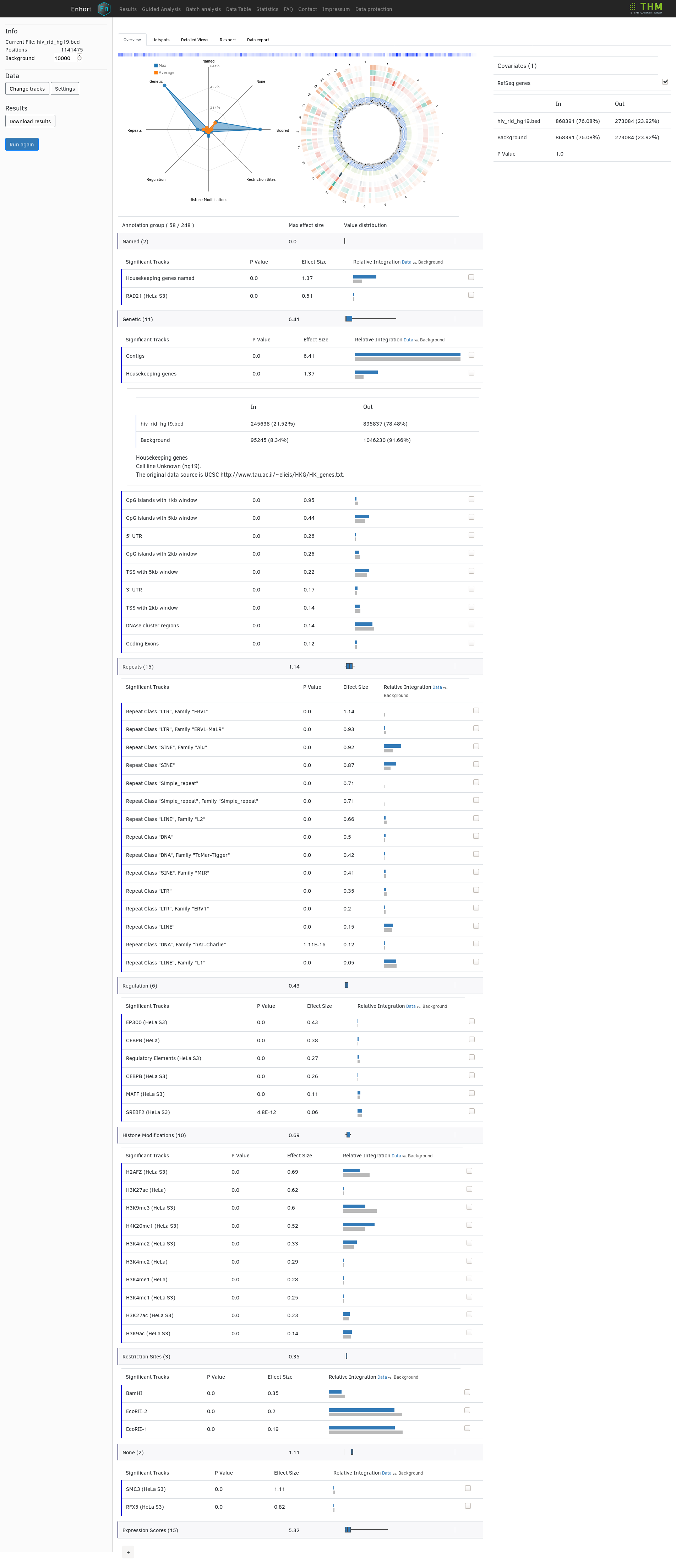
Further possibilites
In the upper part different tabs are located containing further analysis of the given sites, as well as the option to download the background sites or results. Depending on the individual scientific question other covariates can be selected to build a specialized background model.